ABSTRACT:
Objetive: Description of the different isolated microorganisms and their prevalence in infections associated with health care, in addition to determining their patterns of resistance to antibiotics in patients admitted with a confirmed or suspected diagnosis of COVID-19 in the Intensive Care Unit, during a third-level medical center with hospital reconversion.
Method: Patient demographic data was obtained from the clinical record, with defined criteria. Antibiotic resistance patterns were evaluated as well as the identification of isolated bacteria in cultures of expectoration, pleural fluid, catheter tips. For bacterial identification and resistance mechanisms, automated equipment and phenotypic tests were used, following the CLSI (Clinical & Laboratory Standards Institute) criteria.
Results: A total of 100 patients with bacterial infection added to the main COVID-19 picture were obtained, representing pneumonia, urinary tract infection, catheter infections and bacteremia. A total of 100 strains were isolated, of which 84 are Extremely Drug Resistant, 12 Multidrug Resistant and only 4 variable sensitivity. The bacteria with the highest prevalence is Staphylococcus aureus with, followed by Pseudonomas aeruginosa and Stenotrophomonas maltophilia. 100% of the patients admitted to the ICU (Intensive Care Unit) had death.
Conclusion: The increase in resistance to antibiotics in the COVID-19 pandemic has set off alarms due to the complication that this brings, and the improper use of drugs as prophylaxis or attempted treatment only generates selective pressure that leads to an increase in resistance as observed in the isolated strains in this study, where the vast majority present enzymes as well as other resistance mechanisms that confer them to be XDR (Extremely Drug Resistant).
Keywords: antibiotic resistance; COVID-19 disease; nosocomial infections
RESUMEN:
Objetivo: Descripción de los diferentes microorganismos aislados y su prevalencia en infecciones asociadas a la atención de la salud, además de determinar sus patrones de resistencia a antibióticos en pacientes ingresados con diagnóstico confirmado o sospechado de COVID-19 en la Unidad de Cuidados Intensivos, en Centro médico de tercer nivel con reconversión hospitalaria.
Método: Los datos demográficos de los pacientes se obtuvieron de la historia clínica, con criterios definidos. Se evaluaron patrones de resistencia a antibióticos, así como la identificación de bacterias aisladas en cultivos de expectoración, líquido pleural, puntas de catéter. Para la identificación bacteriana y los mecanismos de resistencia se utilizaron equipos automatizados y pruebas fenotípicas, siguiendo los criterios del CLSI (Clinical & Laboratory Standards Institute).
Resultados: Se estudió un total de 100 pacientes con infección bacteriana sumado al cuadro principal de COVID-19, de los cuales representó neumonía, infección de vías urinarias, infecciones de catéter y bacteriemia. Se aislaron un total de 100 cepas, de las cuales 84 son Extremadamente Resistentes, 12 Multirresistentes y solo 4 de sensibilidad variable. La bacteria con mayor prevalencia es Staphylococcus aureus, seguida de Pseudonomas aeruginosa y Stenotrophomonas maltophilia. El 100% de los pacientes ingresados en UCI (Unidad de Cuidados Intensivos) tuvieron muerte.
Conclusión: El aumento de las resistencias a los antibióticos en la pandemia de COVID-19 ha hecho saltar las alarmas por la complicación que esto trae consigo, y el uso inadecuado de fármacos como profilaxis o intento de tratamiento solo genera una presión selectiva que conduce a un aumento de las resistencias como se observa en las cepas aisladas en este estudio, donde la gran mayoría presenta enzimas así como otros mecanismos de resistencia que les confieren ser XDR (Extremadamente Resistente).
Palabras Clave: resistencia a los antibióticos; Covid-19; infecciones nosocomiales
1. INTRODUCCIÓN
In December 2019, a series of novel coronavirus-related pneumonia cases were reported in Wuhan, China. The disease was designed as coronavirus disease 2019 (COVID-19) by World Health Organization (WHO) (1). Genetic sequencing of the virus suggests that it is a betacoronavirus closely linked to the SARS virus (2).
Bacterial coinfections, well documented in other respiratory viral infections, notably infuenza, have not yet been thoroughly studied for COVID-19 (3). While antibiotics are ineffective for treatment of COVID-19, they are prescribed in patients with suspected or documented COVID-19 for a variety of reasons. This includes difficulty in ruling out bacterial co-infection on presentation, but also the possibility of bacterial secondary infection during the course of the illness (4). The prevalence of bacterial infection ranged from 14% to 100% in intensive care unit (ICU) patients. A recent meta-analysis suggested that only 7% of COVID-19 patients were expected to have bacterial infection. However, it should be noted that most patients received antibiotics before they were diagnosed with bacterial infection (5).
According to existing reports, 50% of COVID-19 deaths experienced secondary bacterial infections (SBIs); thus, patients with SBIs have a higher risk of mortality. SBIs had become the hidden threat lurking behind COVID-19. The effective antimicrobial regimen is still one of the key measures for the successful treatment of COVID-19 (6).
Ventilator-associated pneumonia (VAP) is considered one of the most common hospital-acquired infections and the leading infectious cause of mortality in intensive care units (7). Secondary bacterial pneumonia in a patient on invasive mechanical ventilation has a presentation similar to that of hospital-acquired pneumonia but warrants aggressive use of empiric broad-spectrum antibiotics with coverage for Meticillin Resistant Staphylococcus aureus (MRSA), Pseudomonas aeruginosa, and possibly other multidrug-resistant organisms in accordance with the guidelines (8). The mortality is more significant in severe cases compared with the nonsevere group due to the higher coinfection rate in severe patients. For more severely ill patients, they are more likely to receive treatment with invasive catheters, resulting in increased sensitivity to secondary infections with multidrug-resistant pathogens such as Acinetobacter baumannii, Escherichia coli, Pseudomonas aeruginosa, and Enterococcus spp. (9).
Empiric antibiotic therapy should be de-escalated on the basis of microbiology results and clinical judgment, regularly reviewing the possibility of switching of intravenous to oral route of administration and provide targeted treatment based on microbiologic results. Carbapenems have been the backbone for the treatment of these infections, but the development of strains resistant to these antimicrobials raises concerns regarding their use. Antibiotic overuse increases the risk of emergence and transmission of multidrug-resistant bacteria. Infections with multidrug-resistant bacteria are more difficult to treat, and associated with increased morbidity and mortality (7).
The negative impact of the use of antibiotics and antimicrobials in general is still unknown with certainty, but a significant increase in the levels of resistance to them in infections associated with health care is estimated
In this study, our objective was to describe the rate of in-hospital infections and the susceptibility patterns of the causative organisms during the COVID-19 pandemic in patients admitted to the Intensive Care Unit.
2. METHODS AND MATERIALS
A retrospective descriptive cohort study was conducted in a tertiary medical center that underwent hospital reconversion for COVID-19 care. The reorganization of the hospital included the expansion of the Intensive Care Unit (ICU). All consecutive patients admitted with confirmed polymerase chain reaction (PCR) or awaiting sample collection, with severe COVID-19 acquired in the community between the months of October 2020 and January 2021 were included. Defined as severe by the presence of any of the following: respiratory rate ≥ 30 breaths per minute, (Oxygen Saturation) SpO2 <80%, involvement of 50 % or more of the lung parenchyma as evidenced Chest computed tomography or Corads 4 and 5 criteria. On clinical presentation, demographics, laboratory results, antibiotic prescription upon admission and duration, development and characteristics of intrahospital infections and discharge status were obtained from the electronic medical record of each one of the patients.
The primary outcome was the development of a health care associated infection (HAI). For each case of HAI, the causative microorganism and its susceptibility or resistance pattern were recorded. Secondary outcomes included death during hospitalization and length of stay. Hospital-acquired infections were identified and defined according to the standardized criteria of the National Health Security Network (NHSN).
For detection of SARS-Cov-2 by PCR
To collect the sample for detection of SARS-Cov-2, an oropharyngeal and nasopharyngeal swab was performed in which cases it was possible to do so, and nasopharyngeal swab for cases that required intubation, prior to sample collection. They were stored and transported in universal transport medium for viruses and sent to the State Public Health Laboratory for the detection of the pathogen. The same entity that reports the results as positive or negative, according to the presence or absence of viral genetic material.
Bacterial growth and susceptibility testing
Bronchial secretions were obtained by the tracheal aspiration technique in sterile bronchial aspiration traps, by the medical or nursing staff.
The bronchial secretions were analyzed in the clinical laboratory, in the microbiology area. They were processed in a biosafety cabinet, spreads of the sample were prepared on a slide to perform the Gram stain, for the acceptance of the sample with analytical quality, the Murray-Washington criteria were applied,(10) which establish the acceptable quality for culture of the sample, if it has greater than 25 leukocytes per field and less than (10) squamous epithelial cells per field in the observation with the objective of 100X, samples with greater than or equal to (10) epithelial cells per field in 100X were rejected.
The culture for the rest of the samples was carried out by cross-streaking in 5% sheep’s blood agar agar, chocolate agar agar and MacConkey agar, they were incubated at 36 + 1 ºC under microaerophilic conditions for 24 to 48 hours.
Bacterial identification was carried out using panel NC86 and PC33, for gram negative bacilli and for gram positive cocci, respectively. The identification and antibiogram of the microorganism was carried out in a commercial MicroScan4® system (Beckman, USA). The interpretation of the minimum inhibitory concentration in terms of sensitivity was used by the reference criteria of the CLSI (Clinical & Laboratory Standards Institute).
For the detection of Extended Spectrum Betalactamases reported by the team, a phenotypic manual test was performed using the double disc technique (11). On the other hand, for the detection of resistance to methicillin in the case of Staphylococcus, the diffusion test of 1µg oxacillin discs was used, as well as the 30 µg cefoxitin screening (12,13).
The strains of Pseudomonas aeruginosa and Stenotrophomonas were considered XDR, MDR according to the classification issued to the amount of drugs and drug families to which they show resistance (14).
Statistic analysis
Categorical variables were reported as frequencies and proportions. Quantitative variables were reported using mean and standard deviation, or median and interquartile range (IQR) according to their observed distribution. Logistic regression analysis to identify the factors associated with the susceptibility of acquiting bacterial infection.
3. RESULTS
In the following study, it was detected that all patients admitted to the Intensive Care Unit received antibacterial therapy as prophylaxis, indicating third-generation cephalosporins and fluoroquinolones by the medical area. 100% of the study sample received levofloxacin and third-generation cephalosporins simultaneously, prior to bacterial infection or during the course of aggregate infection.
During the study period, 150 patients were admitted to the intensive care unit, but those who showed an aggregate bacterial infection related to health care or nosocomial were taken into account for this study.
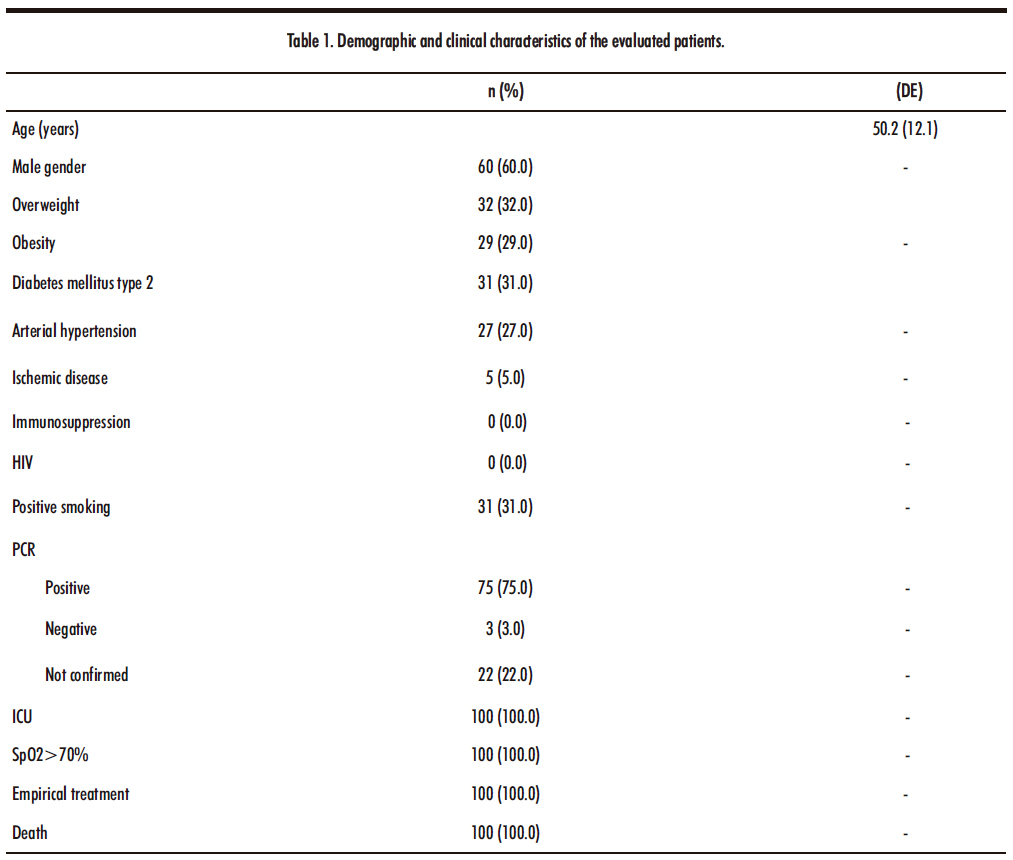
The demographic data and main characteristics of the patients under study (n=100) are reported in Table Number 1. Under the criteria established for data analysis.
Where 60% of the sample corresponds to the male gender, 40% to the female gender. Both genders with 100% death after their hospital stay in the Intensive Care Unit. The average age of the study population is 50.2, with standard deviation of 12.1464.
HIV: Human Immunodeficiency Virus, PCR: Polymerase Chain Reaction, ICU: Intensive Care Unit, SpO2: Oxygen Saturation. Empirical treatment; Levofloxacin and third-generation cephalosporins
75% of the patients admitted to the Intensive Care Unit report a positive PCR test, while 22% the result is pending by the State Public Health laboratory and only 3% have a negative test. However, 100% presented after their stay an aggregated bacterial infection independent of the result for the disease due to covid-19.
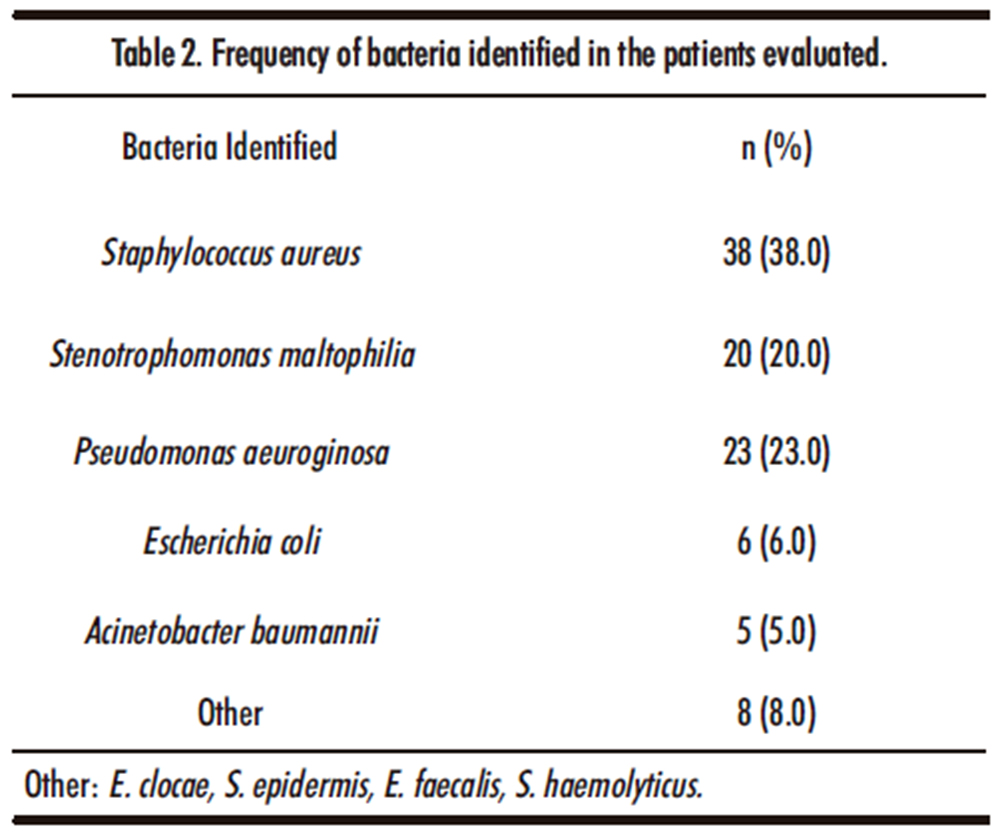
Within the biological samples analyzed that comprise 4 6% bronchial secretion, 32% expectoration, 6% catheter tips, 9% blood culture, 4% urine culture and 2% pleural fluid, the main isolated bacteria are listed in table number 2. With a high incidence of Staphylococcus aureus 38% and Pseudomonas aeruginosa 23% Gram negative bacteria are those that were isolated in the greatest number with 55% and 45% for Gram positive bacteria. Highlighting the absence of fungal development such as Candida spp or Aspergillus spp.
Other: E. clocae, S. epidermis, E. faecalis, S. haemolyticus.
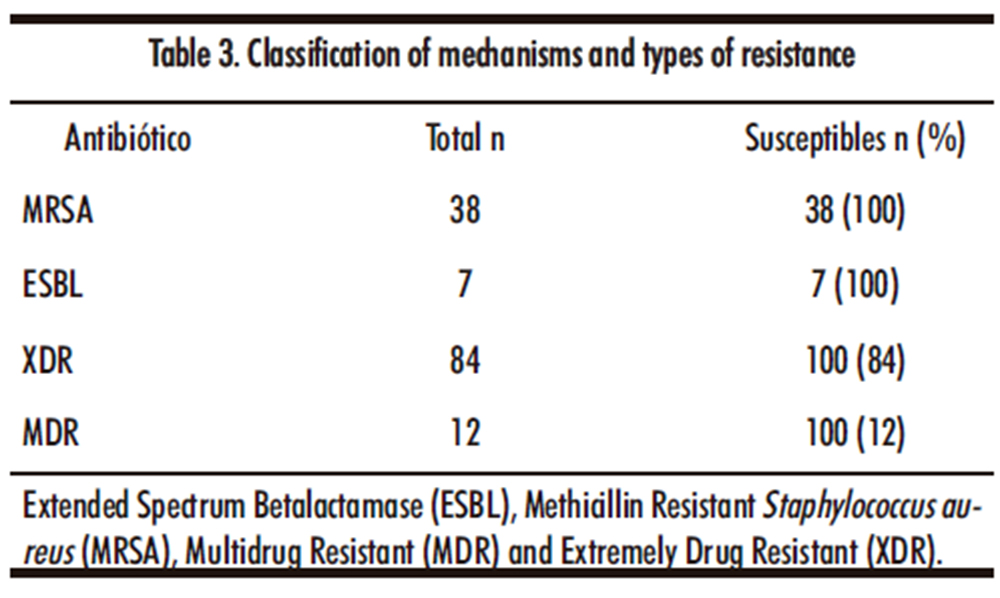
According to the results obtained by the antibiograms and the classification mentioned in the methodology as Extended Spectrum Betalactamase producing bacteria (ESBL), Methicillin Resistant Staphylococcus aureus (MRSA), Multidrug Resistant Bacteria and Extremely Drug Resistant Bacteria are reported in the table number 3. With 84% of Extremely drug resistant and 12% of Multidrug resistant, including those bacteria resistant to methicillin or producing ESBL.
Extended Spectrum Betalactamase (ESBL), Methicillin Resistant Staphylococcus aureus (MRSA), Multidrug Resistant (MDR) and Extremely Drug Resistant (XDR).
According to the results of the antibiograms issued by the microbiology department using MICs issued by the CLSI, a significant number of strains with high resistance to antibiotics are reported, as shown in the table. Where antibiotics are categorized as beta-lactams; penicillin and its derivatives, cephalosporins and carbapenems, the floroquinolones; levofloxacin, ciprofloxacin, mofloxacin, the aminoglycosides; amikacin and gentamicin, tetracyclines; tetracycline and tigecycline, and some other drugs with different classifications, such as glycopeptides, macrolides, lincosamines, among others.
Table No. 4 shows the total number of isolates of said bacteria as n and the % of them that show resistance to the indicated antibiotic.
A logistic regression analysis was carried out to identify the factors that are associated with the susceptibility to the development of a bacterial infection. Prior to the analysis, two CNCD (Chronic Disease Prevention and control) variables were created, as well as a nutritional diagnosis.
4. DISCUSSION
We conducted a retrospective descriptive study to describe the number of health care-associated infections and the resistance patterns of isolated microorganisms in patients with severe COVID-19 admitted to the Intensive Care Unit at a tertiary medical center in the State of Mexico. Generalized empirical antimicrobial prescriptions of beta-lactams as third-generation cephalosporins were found, among which ceftazidime and ceftriaxone stood out, as well as a treatment with the fluoroquinolone levofloxacin, both antibacterial treatments administered simultaneously from admission to the ICU. In our study, initially 100% of the study patients and those who did not enter the study due to exclusion criteria had the aforementioned prophylactic antimicrobial therapy, something that contrasts with a study carried out by Vaughn et al., Who reported the use of empirical antibiotics in 56.6% of COVID-19 patients admitted to 38 Michigan hospitals. In that study, empirical use of antibiotics ranged from 27% to 84% among hospitals, and the most frequently prescribed antibiotic was ceftriaxone in 38.9%. Although the prevalence of bacterial coinfections on admission is low (2.4-7%), empirical antimicrobial treatment is extremely common (72-90%) (15). Taking into account the amount of bacterial coinfections in patients with the disease of the COVID-19 so low, the World Health Organization suggested not using antibiotics to treat the disease (16).
On the other hand, the use of levofloxacin is justified as an initial treatment for nosocomial pneumonia or community-acquired pneumonia due to its wide spectrum and good concentration achieved in the lungs,(17) as well as the use of third-generation cephalosporins such as ceftriaxone at a dose of 1 and 2 g.(18). However, when presenting a co-infection of bacterial etiology, I modify the treatment to a targeted therapy in most cases, this prophylactic treatment was used for 7 to 10 days according to the results of the cultures performed., where antimicrobial therapy had to be scaled to carbapenemics, colistin, vancomycin or linezolid, according to the isolated microorganisms.
However, resistance to antibiotics suggests that bacteria already have resistance to most of the drugs of each of the antimicrobial families (19) and that it gives them a classification of Extremely Drug Resistant, of which there are 84 strains and Multi Drug Resistant with 12 strains.
In a study by Li J et al. A total of 159 strains of bacteria were isolated from the cultures in the 102 patients. Among the isolated bacteria, Gram negative bacteria were the main bacteria, representing 85.5%. The three main bacteria were Acinetobacter baumannii (A. baumannii, 35.8%), Klebsiella pneumoniae (K. pneumoniae, 30.8%), and Stenotrophomonas maltophilia (S. maltophilia, 6.3%) (20). While in our study, the Gram positive bacterium S. aureus represents a total of 38% of the isolates and 100% in resistance to methicillin by the oxacillin test and the cefoxitin test. On the other hand, in Gram negative isolates, P. aeruginosa with 23% and S. maltophilia with 20% of the total of the isolates were the ones that had the highest frequency in the total of samples. A baumannii, which in Li’s study was the most isolated bacterium in our study, represents only 5% of the isolates.
Only 79% of bacteria isolated by co-infection correspond to pneumonia associated with mechanical ventilation or associated with health care, while 21% correspond to infections of the pleural fluid, catheter infections, bacteremia and urinary tract infections.
A high number of strains resistant to beta-lactams are observed, both for penicillin and its derivatives with 100%, penicillin derivatives with beta-lactamase inhibitor such as ampicillin / sulbactam, amoxicillin / clavulanic acid, piperacillin / tazobactam, also with a resistance of 100%. In the case of first, second, third and fourth generation cephalosporins, 100% resistance is observed, except for ceftazidime, which presents resistance of only 96.3% thanks to the susceptible strains of S. maltophilia. For ESBL-producing strains, high resistance to penicillins and cephalosporins is expected due to their enzymatic activity (21), for the case of carbapenemase-producing strains they also explain the high resistance index to beta-lactams (22) already mentioned as well as the carbapenemics meropenem, ertapenem and imipinem resistance of 83.9%, 93.5% and 89.7% respectively. The strains with the highest production of carbapenemases in this study are P. aeruginosa XDR and A. baumannii XDR. In the case of high levels of resistance to all beta-lactams for S. aureus, it is explained by resistance to methicillin, which implies not an enzyme but the modification of the PBP Penicillin-Binding Proteins that confers resistance to all beta-lactams (23).
Quinolone resistance is caused by various mechanisms, particularly plasmid-ediated quinolone resistance (PMQR) which contains the pentapeptide repeat family Qnr proteins (QnrA, QnrB, QnrS, QnrC, and QnrD). These proteins confer quinolone resistance by physically protecting DNA gyrase and topoisomerase IV from quinolone acts. This condition may provide a selective advantage for the development of quinolone resistance which could result in therapeutic failure (24). Quinolone resistance is more common in Enterobacteriaceae that show resistance to beta-lactams due to the production of extended-spectrum beta-lactamases (ESBL), acquired class C beta-lactamases or carbapenemases (25).
A study carried out by López-Jacome and collaborators carried out in the second half of 2019 and the second half of 2020 in 35 health centers throughout Mexico, compares the resistance acquired by gram positives and gram negatives, finding resistance of up to 100 in S. aureus resistant to oxacillin, as well as up to 50% increase in resistance to erythromycin and clindamycin. As well as resistance patterns similar to carbapenems and fluoroquinolones by gram negatives and non-fermenters (26).
One of the great limitations in the study was not having the means to carry out a detailed characterization at the molecular level of the different carbapenemases expressed by the isolated MDR and XDR organisms, as well as the ESBL of the producing strains, which would allow us to know the mechanisms exact resistance of the antibiotics used in the antibiogram and give a correct directed therapy.
Another limiting point of the present study is the lack of manual and automated phenotypic and genotypic tests for the detection of colistin resistance, for which reason the absence of PDR-resistant Pan Drug-resistant bacteria cannot be assured, since they were not considered in the present study. But some of the Pseudomonas aeruginosa and Acinetobacter baumannii cultures after colistin treatment persisted in subsequent control cultures. Which indicates possible mechanisms of resistance to the last line of treatment.
In this study, the targeted therapies used for the treatment of isolated bacteria are not mentioned, due to a later study that is intended to report to indicate resistance patterns in late-line drugs for the isolates obtained and the therapeutic successes or failures, in addition to the considerations that are taken into account in the choice of medication.
As an analysis of the logistic regression we have the following. Finding that having a previous Chronic Disease Prevention and control (CNCD) such as type 2 diabetes mellitus, hypertension or ischemic disease increases the risk by 10,345 (IC 1,127-94,995) (times more likely to develop E. Coli.)
On the other hand, analyzing the factors that are associated for the development of infection by A. baumannii, being a man increases by 19,650 (IC 1,187-325,389) the probability of acquiring the disease compared to being a woman, as well as having positive smoking increases the risk in 31,582 (CI 1,469-679,621) times with respect to those who do not smoke, being statistically significant in both cases.
For the case of other bacteria (E. clocae, S. epidermidis, S. haemolyticus, E. faecalis) the body weight classified as normal weight, overweight and obesity was analyzed. Finding that having a normal weight reduces the risk of developing a bacterial pathology by 12,345 (IC 0.009-0.760) compared to those who are overweight or obese.
5. CONCLUSION
The use of prophylactic antibacterial treatment with fluoroquinolones and cephalosporins did not represent an advantage, since the patients acquired a bacterial coinfection where the strains are resistant to these drugs.
Knowing the local epidemiology is of vital importance to be able to determine the treatments and suspicions of etiological agents of associated infections and to prescribe an empirical treatment as close to the directed one as possible.
Nosocomial infections are a problem that is leading to the death of people in serious health conditions in the Intensive Care Units.
The increase in resistance to antibiotics in the COVID-19 pandemic has set off alarms due to the complication that this brings, and the improper use of drugs as prophylaxis or attempted treatment only generates selective pressure that leads to an increase in resistance as observed in the isolated strains in this study, where the vast majority present enzymes as well as other resistance mechanisms that confer them to be XDR.
There was an increase in the number of health care-associated infections, and bacteria showed greater resistance to bacteria isolated in similar pre-pandemic infections. Hospital reconversion and the pandemic appear to be an important driver of resistance.
Ethical approval. The study design is retrospective, so no intervention or intentional modification is made in the physiological, psychological or social variables in the individuals participating in the study, so the ethics committee did not require registration, but it did approve its development, and due to the aforementioned characteristics, informed consent was omitted.
Acknowledgment
We thank the institutions that supported in carrying out this research.
6. REFERENCES
1. Ma Y, Zeng H, Zhan Z, et al., 2020, Corticosteroid Use in the Treatment of COVID-19: A Multicenter Retrospective Study in Hunan, China., Front. Pharmacol., 11, 1198 https://doi.org/10.3389/fphar.2020.01198
2. Cisneros J, Rosso-Fernández M, Roca-Oporto C. et al. 2019, Colistin versus meropenem in the empirical treatment of ventilator-associated pneumonia (Magic Bullet study): an investigator-driven, open-label, randomized, noninferiority controlled trial. Crit Care., 23, 383 https://doi.org/10.1186/s13054-019-2627-y
3. Kreitmann L, Monard C, Dauwalder O, Simon M, Argaud L, 2020, Early bacterial co-infection in ARDS related to COVID-19, Intensive Care Med., 46, 1787-1789 https://doi.org/10.1007/s00134-020-06165-5
4. Langford B, So M, Raybardhan S, et al. 2020, Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect., 26(12), 1622-1629. doi:10.1016/j.cmi.2020.07.016
5. Liu C, Wen Y, Wan W, Lei J, Jiang X., 2021, Clinical characteristics and antibiotics treatment in suspected bacterial infection patients with COVID-19, Int. Immunopharmacol., 90, 107157 https://doi.org/10.1016/j.intimp.2020.107157
6. Li, J., Wang, J., Yang, Y. et al. 2020, Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: a retrospective analysis. Antimicrob Resist Infect Control, 9, 153 https://doi.org/10.1186/s13756-020-00819-1.
7. WHO. COVID-19 Clinical management: living guidance. OMS / 2019-nCoV / Clinical / 2021.1, 85 https://www.who.int/publications/i/item/WHO-2019-nCoV-clinical-2021-1
8. Wu C, Adhi F, Highland K.,2020, Recognition and management of respiratory co-infection and secondary bacterial pneumonia in patients with COVID-19, Clevel. Clin. J. Med., 87(11), 659-663 doi: https://doi.org/10.3949/ccjm.87a.ccc015
9. Chen, X., Liao, B., Cheng, L. et al., 2020, The microbial coinfection in COVID-19. Appl Microbiol Biotechnol, 104, 7777-7785 https://doi.org/10.1007/s00253-020-10814-6
10. Murray P, Washington J. 1975, Microscopic and bacteriologic analysis of expectorated sputum. Mayo Clin Proc. 50, 339,344. https://doi.org/10.1080/00357529.1975.11762852
11. Babic M, Hujer A, Bonomo R., 2006, What´s new in antibiotic resistance? Focus on beta-lactamases, Drug Resist Updat., 9(3), 142-56 doi: 10.1016/j.drup.2006.05.005
12. Mathews A, Thomas M, Appalaraju B, Jayalakshmi J., 2012, Evaluation and comparison of tests to detect methicillin resistant S. aureus., Indian J Pathol Microbiol., 53, 79-82
13. Louie L, Matsumura S, Choi E, et al., 2000, Evaluation of Three Rapid Methods for Detection of Methicillin Resistance in Staphylococcus aureus, J.Clin. Microbiol.,38(6),2170-2173
14. Yuan X, Liu T, Wu D, Wan Q., 2018, Epidemiology, susceptibility, and risk factors for acquisition of MDR/XDR Gram-negative bacteria among kidney transplant recipients with urinary tract infections., Infect Drug Resist., 11, 707-715 doi: 10.2147/IDR.S163979
15. Martínez-Guerra B, González-Lara M, de-León-Cividanes N, et al,.2021, Antimicrobial Resistance Patterns and Antibiotic Use during Hospital Conversion in the COVID-19 Pandemic. Antibiotics., 10(2), 182 https://doi.org/10.3390/antibiotics10020182
16. Getahun H, Smith I, Trivedi K, et al., 2020, Tackling antimicrobial resistance in the COVID-19 pandemic. Bull World Health Organ., 98, 442-442A doi: http://dx.doi.org/10.2471/BLT.20.268573
17. Shorr A, Zadeikis N, Jackson W, Ramage A, Wu S, Tennenberg A, Kollef M., 2005, Levofloxacin for treatment of ventilator-associated pneumonia: a subgroup analysis from a randomized trial. Clin Infect Dis., 15, 40 Suppl 2:S123-9. doi: 10.1086/426192. PMID: 15712100.
18. Telles J, Cieslinski J, Gasparetto J, Tuon F., 2019, Efficacy of Ceftriaxone 1 g daily Versus 2 g daily for The Treatment of Community-Acquired Pneumonia: A Systematic Review with Meta-Analysis, 17:7, 501-510, doi: 10.1080/14787210.2019.1627872
19. Arroyo-Mejía M, Valdez-Arcos O, Almeida-Villegas JA, 2019, Resistance to antibiotics; a perspective, Rev. Med. Inv., 7(2), 67-72
20. Li, J., Wang, J., Yang, Y. et al. 2020, Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: a retrospective analysis. Antimicrob Resist Infect Control 9, 153. https://doi.org/10.1186/s13756-020-00819-1
21. Almeida-Villegas JA, Rodríguez M, Rodríguez S, Gutiérrez-Gutiérrez J.A., 2020, Resistance to Antibiotics in Clinical Isolates of Extended-Spectrum BetaLactamase (ESBL)-Producing Escherichia coli Obtained from Urine Cultures in Patients with Urinary Tract Infection, Acta Microbiol Bulg., 36 (3), 100-103
22. Patel J, Rasheed K, Kitchel B,. 2009, Carbapenemases in Enterobacteriaceae: Activity, Epidemiology, and Laboratory Detection, Clin. Microbiol. Newsl., 31(8), 55-62 https://doi.org/10.1016/j.clinmicnews.2009.03.005
23. Pantosti A, Sanchini A, Monaco M, 2007, Mechanisms of antibiotic resistance in Staphylococcus aureus, Future Microbiol, 2(3), 323-334 https://doi.org/10.2217/17460913.2.3.323
24. Almeida-Villegas J, Carrillo I, Garcia-Contreras R, Peña S, Quinolone resistance in different uropathogens isolated from urine cultures in patients with urinary tract infection, bioRxiv 2020.09.07.286674; doi: https://doi.org/10.1101/2020.09.07.286674
25. Machucaa J., Agüero J., Miro E., Conejo M. C., Oteo, J., et al., 2017, Prevalence of quinolone resistance mechanisms in Enterobacteriaceae producing acquired AmpC b-lactamases and/or carbapenemases in Spain. Enferm Infecc Microbiol Clin., 35(8), 487-492 https://doi.org/10.1016/j.eimc.2016.05.006
26. López-Jacome E., et al., 2022, Increment Antimicrobial Resistance During the COVID-19 Pandemic: Results from the Invifar Network., Microb Drug Resist., 28(3),338-345. doi: 10.1089/mdr.2021.0231.